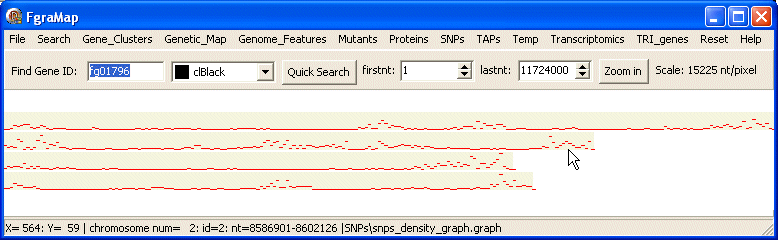
graph data
This is used to display a graph or histogram along a chromosome, e.g. the SNP density using the count of SNPs/50,000 nt. The value of the last field on a data line is used to determine the y-value on the graph.
This is part of the data file for snps_density_graph.graph:
# clRed # min 0 max 151 # chrom start end SNPs 1 1 50000 1 1 50001 100000 0 1 100001 150000 11 1 150001 200000 41 1 200001 250000 31 1 250001 300000 36 1 300001 350000 50 1 350001 400000 50
This shows the SNP density as a graph or histogram.
The first #-line is essential and contains the colour used to draw the graph.
The second #-line is not essential but contains the max and min values used to draw the y-axis of the graph. If this is not present OmniMapFree finds these values by checking all the data.
The third line is blank and is not essential but separates the #-line from the data lines.
The following lines contain the data for each feature displayed. The first three data fields: chromosome id (chrom), start position (start) and end position (end) are used by OmniMapFree for the x-axis.
The last data field contains the value used for the y-axis.
The max value provided on the second #-line can be used to change the scale of the graph. The top of a chromosome is set to the max y-value - this makes it easier to see variations in the lower data values. Any data values greater than the "max" are reduced to the max value and are drawn as a black line to indicate that they have been truncated.
Here is the map drawn by the snps_density_graph.graph data file: