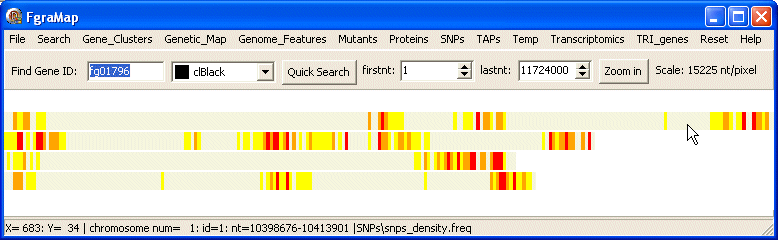
freq data
The freq data file is used to display chromosome regions in different colours. The colour is determined by the value of the last field and a colour look-up list in the first #-line.
This is part of the data file for snps_density.freq:
#clBeige 18 clYellow 38 clOrange 69 clRed #chrom start end SNPs 1 1 50000 1 1 50001 100000 0 1 100001 150000 11 1 150001 200000 41 1 200001 250000 31 1 250001 300000 36 1 300001 350000 50 1 350001 400000 50
The first #-line contains a tab-delimited list of colour and cutoff values. Up to 20 different colours can be used so users can create colour gradients to represent gene density, %GC, recombination frequency, etc.
In this case any last field value less than 18 is clBeige, from 18 to less than 38 is clYellow, from 38 to less than 69 is clOrange, and anything 69 or greater is clRed.
The second #-line is not essential but is used to describe the different columns/fields.
The third line is blank and is not essential but separates the #-line from the data lines.
The following lines contain the data for each feature displayed. The first three data fields: chromosome id (chrom), start position (start) and end position (end) are used by OmniMapFree to draw the feature.
The last data field contains the value used to determine the colour from the look-up list in the first #-line.
This freq data file draws a similar genome map to the one from the expr data file - but in a different way.
In this case the features are 50,000 nt regions of the chromosome, the fourth and last field shows the number of SNPs in this particular region, and is used to look up the colour. The advantage of the freq data file over the expr data file is that the colours are specified on one line - the first #-line so are more easily changed.
Here is the map drawn by the snps_density.freq data file: