posn data
The posn data file is used to draw genes or features based on their position in the genome (i.e. chromosome id and position on the chromosome).
This is part of the data file for TRI_genes.posn:
# clBlue # chrom start end strand name annotation 1 282078 283832 + fg00071 TRI1 cytochrome P450 monooxygenase 2 5384739 5386074 - fg03532 TRI8 C-3 esterase 2 5388953 5390718 + fg03534 TRI3 15-O-acetyltransferase 2 5391676 5393415 - fg03535 TRI4 P450 C-2 monooxygenase 2 5394569 5395239 + fg03536 TRI6 Zinc finger transcription factor 2 5398128 5399312 + fg03537 TRI5 Sesquiterpene cyclase
The first #-line contains the colour used to display the genes or regions. Users can put different genes in different posn data files and have them drawn in different colours.
The second #-line is not essential but describes the different columns.
The third line is blank and is not essential but separates the #-lines from the data lines.
The following lines contain the data for each gene displayed. The first three data fields: chromosome id (chrom), start position (start) and end position (end) are used by OmniMapFree to draw the gene.
The remaining data fields: strand, gene id (name) and annotation are also essential but are only used when you display the Gene List Dialog box after selecting a region of the map.
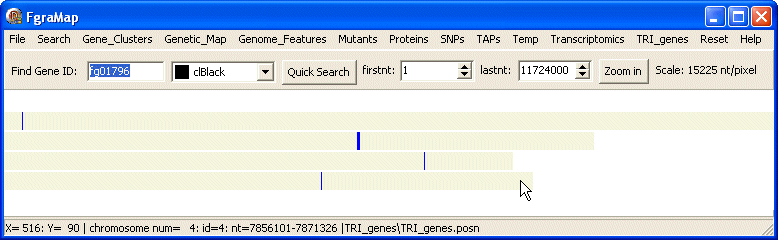
Here is the map drawn by the TRI_genes.posn data file: