expr data
The expr data file is used to display genes or features using different colours. This was originally used for microarray expression data where the induced genes might be coloured green and the repressed genes coloured red. But has many other uses e.g. for proteomics data: proteins present/increasing and absent/decreasing could be shown in different colours.
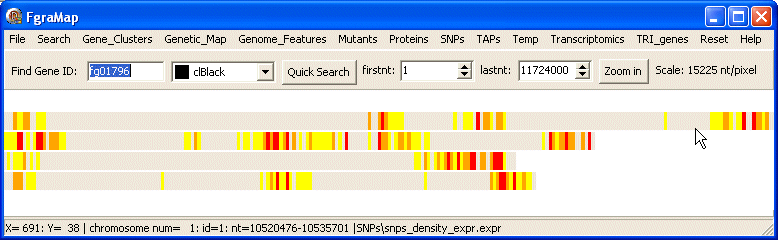
This is part of the data file for snps_density_expr.expr:
#chrom start end SNPs colour 1 1 50000 1 clBtnFace 1 50001 100000 0 clBtnFace 1 100001 150000 11 clBtnFace 1 150001 200000 41 clOrange 1 200001 250000 31 clYellow 1 250001 300000 36 clYellow 1 300001 350000 50 clOrange 1 350001 400000 50 clOrange
The first and only #-line is not essential but is used to describe the different columns/fields.
The second line is blank and is not essential but separates the #-line from the data lines.
The following lines contain the data for each feature displayed. The first three data fields: chromosome id (chrom), start position (start) and end position (end) are used by OmniMapFree to draw the feature.
The last data field contains the colour used to draw the feature.
In this case the features are 50,000 nt regions of the chromosome, the fourth field shows the number of SNPs in this particular region, and was used to determine the colour. But the colour specified in the last field does not have to depend on any other field.
The colours were chosen to produce a colour gradient from gray-yellow-orange-red to indicate increasing SNP density.
Here is the map drawn by the snps_density_expr.expr data file: